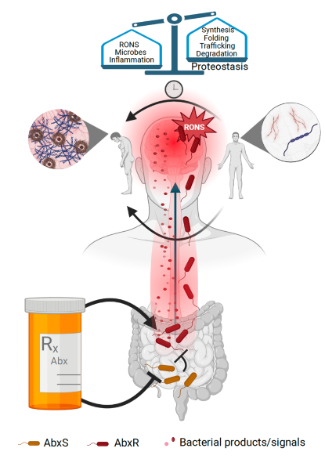
Protein conformational diseases (PCDs) are characterized by a progressive loss of neuronal or muscle function due to protein misfolding and aggregation, a common feature among diseases such as Alzheimer’s, Parkinson’s, Huntington’s, or Lou Gehrig’s diseases (ALS). The exact factors that influence PCDs are not known. Recent evidence suggests that bacteria may contribute to the pathogenesis of these neurodegenerative diseases. To better understand the influence of bacteria on protein homeostasis (proteostasis), we are studying the effect of bacterial colonization of the Caenorhabditis elegans gut on protein aggregation in the intestine and other tissues. We found two gram-negative species, Pseudomonas aeruginosa and Klebsiella pneumoniae, that enhanced protein aggregation in the intestine nearly five-fold; these two strains also affect proteostasis across other tissues, including muscle and neurons. Both species can be found as part of the normal human microbiome and are known opportunistic pathogens. An increase in the abundance of these bacteria within the human gut was previously linked with the enhanced progression of neurodegenerative diseases. Collectively, these results suggest that intestinal bacteria affect the host folding environment; however, which bacterial factors are responsible for the enhancement of aggregation remains unknown. Moreover, which bacteria within the entire microbiome enhance or suppress host proteostasis is not known. Our results, along with other published data, demonstrate that butyrogenic bacteria and butyrate suppress protein aggregation and the associated toxicity; however, further work is needed to elicit the exact mechanisms. Collectively, we employ C. elegans to reach the following goals:
(i). Identify bacterial genes that disrupt host proteostasis
(ii). Characterize the effect of the human microbiome bacterial isolates on host proteostasis
(iii). Decipher the mechanisms by which butyrate suppresses bacteria-mediated proteotoxicity
The importance of this project lies in its potential to identify microbial signatures and molecules associated with the pathogenicity of PCDs, thus providing opportunities to develop novel diagnostics, prophylactics, and therapeutics.