Daniel Czyz
Associate Professor
-
Teaching Interests
- Antimicrobial Resistance: MCB4271 & MCB5270
- Antimicrobial Resistance Journal Colloquy
- Antimicrobial Resistance Laboratory: MCB4271L
-
Education
- 2005 - University of Illinois, Chicago – BS, Biochemistry
- 2012 - Northwestern University – Ph.D., Biological Sciences/Biotechnology
- 2018 - University of Chicago – Postdoctoral Scholar, Microbiology
-
Description of Research
Dr. Czyz is interested in two topics: antimicrobial resistance (AMR) and protein conformational diseases (PCDs). Although these two areas look quite diverse, they are not. Antibiotics are the major contributor to AMR. Recent research suggests that bacteria within the human microbiome contribute to the pathogenicity of PCDs, including Alzheimer’s, Parkinson’s, Huntington’s or ALS; therefore, the onset and progression of these ailments is likely affected by antibiotic-induced dysbiosis. The research in the Czyz Lab further explores these two areas:
Antimicrobial Resistance
Antibiotic resistance is becoming the center of a global healthcare crisis. It is estimated that by 2050, antibiotic resistance will claim ten million lives annually and exert a projected economic burden of $100 trillion. Antibiotics target bacteria, which under high selective pressure, rapidly develop resistance. The overuse of antibiotics in the healthcare and agricultural industries has fueled irreversible resistance to drugs that were once effective in treating common bacterial infections. In addition, the development of novel antibiotics has almost completely ceased. With increasing antibiotic resistance and limited treatment options, bacterial infections are once again becoming intractable, leaving a disastrous socio-economic footprint on humans. While much emphasis has been placed on combating bacteria directly, we have taken a different approach; namely, by targeting the host, rather than the pathogen. Unlike conventional pathogen-targeting antibiotics, this host-targeting strategy can prevent and/or treat infections by:
(i) Modulating uptake of bacteria by phagocytic host cells
(ii) Intercepting host pathways or nutrients utilized by the pathogen during infection
(iii) Enhancing the ability of host cells to kill bacteria
Augmenting the ability of host cells to clear invading bacteria provides a promising strategy. As such, we hypothesize that the host-targeting approach can circumvent antibiotic resistance and provide a powerful tool in the fight against antibiotic resistance. We employ small molecules to enhance the ability of phagocytic immune cells to scavenge, uptake and kill bacteria, then introduce them into the host either as a prophylactic or therapeutic intervention. Similar cell-based immunotherapy has been successfully applied in cancer treatment, but such a concept has never been explored in the area of infectious diseases. The results of our work are expected to have a positive impact on human health because they will spur the development of new treatments for patients who are predisposed to bacterial infections. Moreover, our results will broaden the current understanding of host-pathogen interaction, which can potentially help in the development of additional therapeutics.
The Czyz Research Group explores additional non-traditional approaches to battle AMR. We have generated a cocktail of bacteriophages with broad spectrum specificity against about 90% of clinical isolates of Pseudomonas aeruginosa. Our laboratory is currently characterizing these phages and exploring their utility in human and animal applications. Among other non-traditional approaches, we work on silver nanoparticles – these metallic particles have potent antimicrobial activity. We explore the antimicrobial mechanisms of silver nanoparticles and their applicability as potential therapeutic.
Dr. Czyz teaches two courses on AMR: lecture and laboratory. He is also actively involved in fighting AMR outside of his laboratory. He is the Chair of the Advisory Council at the National Institute of Antimicrobial Resistance Research and Education (NIAMRRE).
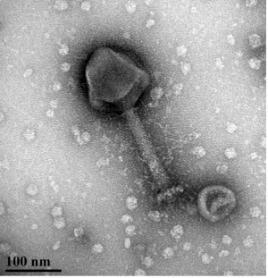
aeruginosa bacteriophage. Credit: Garrett Ellward
Protein Conformational Diseases
PCDs are characterized by a progressive loss of neuronal or muscle function due to protein misfolding and aggregation, a common feature among diseases such as Alzheimer’s, Parkinson’s, Huntington’s or Lou Gehrig’s diseases (ALS). The exact factors that influence PCDs are not known. Recent evidence suggests that bacteria may contribute to the pathogenesis of these neurodegenerative diseases. To better understand the influence of bacteria on protein homeostasis (proteostasis), we are studying the effect of bacterial colonization of the Caenorhabditis elegans gut on protein aggregation in the intestine and other tissues. We found two Gram-negative species, Pseudomonas aeruginosa and Klebsiella pneumoniae, that enhanced protein aggregation in the intestine by nearly five-fold; these two strains also affect proteostasis across other tissues, including muscle and neurons. Both species are part of the normal human microbiome and are known opportunistic pathogens. An increase in the abundance of these bacteria within the human gut was previously linked with the enhanced progression of neurodegenerative diseases. Collectively, these results suggest that intestinal bacteria affect the host folding environment; however, which bacterial factors are responsible for the enhancement of aggregation remains unknown. Moreover, which bacteria within the entire microbiome enhance or suppress host proteostasis is not known. Our results, along with other published data, demonstrate that butyrogenic bacteria and butyrate suppress protein aggregation and the associated toxicity; however, further work is needed to elicited the exact mechanisms. Collectively, we employ C. elegans to reach the following goals:
(i). Identify bacterial genes that disrupt host proteostasis
(ii). Characterize the effect of the human microbiome bacterial isolates on host proteostasis
(iii). Decipher the mechanisms by which butyrate suppresses bacteria-mediated proteotoxicity
The importance of this project lies within its potential to identify microbial signatures and molecules associated with the pathogenicity of PCDs, thus providing opportunities to develop novel diagnostics, prophylactics and therapeutics.
- Publications
-
Awards & Honors
- 2024-2025 UF International Center International Fellow

Contact Information

352-392-0237
Office:
Rm. # 1004
Microbiology Building 981
